
PROTEIN SCAFFOLD MINIMAL SOFTWARE
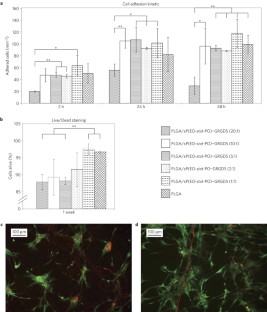
This simple yet versatile model system can help elucidate the molecular interactions implicated in MLOs and pave ways to a new type of biomimetic materials. Batch-enabled Scaffold Software plug-in export/convertor functionality of ProteinLynx Global. Amino acid structural propensities measured in 'host-guest' model studies are often used in protein structure prediction or to choose appropriate residues in de novo protein design. A minimal beta-hairpin peptide scaffold for beta-turn display. Moreover, the droplets formed are capable of recruiting proteins and RNAs and providing a favorable environment for a biochemical reaction with highly enriched components, thereby mimicking the function of natural MLOs. 154012, pMG023 MBP-Axin 384-518-His (miniAxin), Expresses MBP-Axin384-518-His (human Axin minimal scaffold fragment as a fusion protein with MBP and His-. A minimal beta-hairpin peptide scaffold for beta-turn display. Thresholds are set through three dropdowns at the top of the screen, the Protein Threshold, the Min Peptides and the Peptide Threshold. sequence of interest, and (c) display minimal self. Hybrids undergo LLPS into micron-sized liquid droplets resembling LLPS in vitro and in living cells. In a decoy search, you may use probability thresholds, or you may specify FDR targets and allow Scaffold to determine the best settings to achieve them. into a loop of protein L, a mechanically strong and soluble scaffold protein that acts as a display. Here, we report the first design of synthetic hybrids consisting of short oligopeptides of fewer than 10 residues as "stickers" and dextran as a "spacer" to recapitulate the characteristics of IDPs, as exemplified by the multivalent FUS protein. Ferritin molecules, hollow protein spheres that store and release iron as needed, also hold promise for disease diagnosis, drug delivery, and vaccine development. Genetic engineering is generally applied to reconstruct IDPs harboring over 100 amino acid residues. large and spatially proximal paratopes, minimal newly exposed SASA upon terminal trimming, and a. Binding function is evaluated against several molecular targets to determine which proteins evolve specific binding variants. In a decoy search, you may use probability thresholds, or you may specify FDR targets and allow Scaffold to determine the best settings to achieve them.
Liquid-liquid phase separation (LLPS) is an emerging and universal mechanism for intracellular organization, particularly, by forming membraneless organelles (MLOs) hosting intrinsically disordered proteins (IDPs) as scaffolds. Protein scaffold libraries consisting of millions of unique variants are expressed with diversified binding interfaces. Moreover, although bioactive signalling combined with minimization of non-specific protein adsorption is an advanced modification technique for flat surfaces 6, it is usually not accomplished for.


 0 kommentar(er)
0 kommentar(er)
